Scientists of CoEBoC, Tampere University figured out how to acquire accurate 3D images of subcellular structures and functions even from fully opaque samples using X-ray tomography. Their work raises the potential of commercial X-ray microtomography (µCT) devices to a new level for solving problems in biomedical research areas such as in tissue engineering and organ-on-a-chip models.
Light microscopy has multiple good features, but the common problem is the small imaging depth. In addition to limitations of the used optics, the optical properties of the samples can further limit the achievable imaging depth. Even seemingly transparent samples can disturb the transmission of light too much for the acquisition of reliable imaging data. In the worst case, optical images cannot be acquired below the immediate surface, an issue that could leave the internal structures and circumstances in uncertainty.
“It is a big challenge for the development of tissue-engineering biomaterials and organ-on-a-chip platforms if significant parts of these biological entities cannot be assessed”, says doctoral student Ilmari Tamminen, the corresponding author of the recently published paper on Communications Biology.
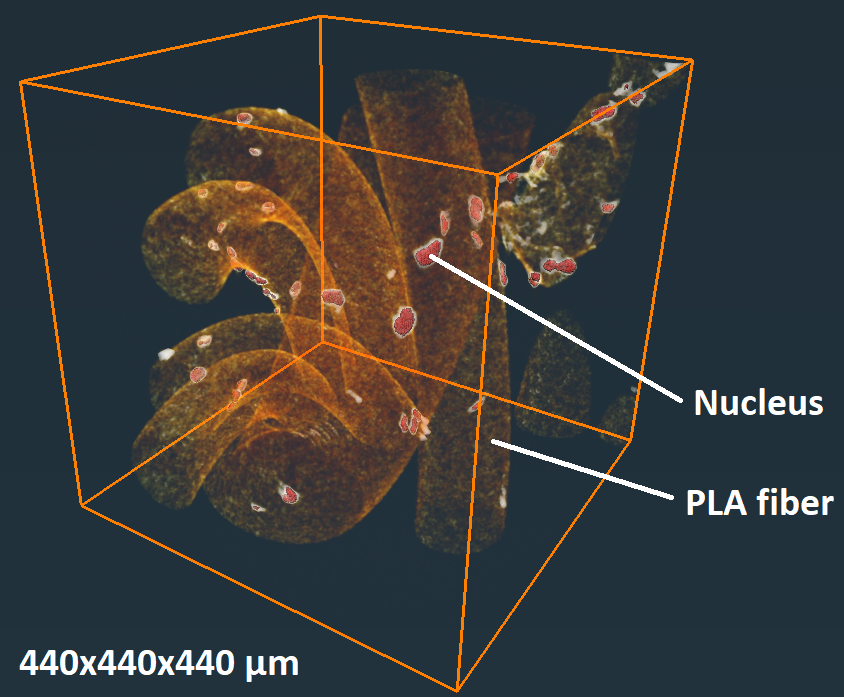
Computational Biophysics and Imaging Group wanted to investigate with their collaborators, could it be possible to accurately image and measure microscopic cell structures with commercial tube-source µCT devices. The principle is similar to clinical computed tomography, but µCT devices can capture more accurate 3D images of smaller specimens. However, these devices cannot image the low-density variations of the subcellular objects as such. To achieve the goal, the scientists combined the antibody-labeling of cell structures, optimized X-ray imaging, and a new data-processing technique. The antibody labeling generates localized nanosilver in the defined biological parts of the sample, attenuating the X-radiation and forming shadows that correspond to the studied cell structures. The attenuating structures can then be located in 3D by X-ray imaging the samples from hundreds of different directions.
“The idea is to optimize the labeling, µCT imaging, and the data processing by the aid of optical reference, then execute the deep 3D imaging using the found parameters which can be used to overcome some of the inaccuracies typical for tube-source based X-ray imaging”, Tamminen describes.
With the new techniques, the scientists were able to accurately 3D image and measure cell structures within clinically-relevant-sized biomaterial constructs in which the conventional microscopy techniques did not reach. The scientists were able to detect microscopic deformations of cell nuclei deep within the constructs after the cells were exposed to a substance affecting the cytoskeleton. In addition to the depth range, the µCT imaging can capture volumes that are relatively large compared to other imaging methods. This enables the acquisition of large samples of cells that make the statistical analyses more reliable.
“I wonder, should this be called microscopy or viewing with a telescope. The optimized µCT imaging of the nuclei corresponds to a situation of defining the inflation state by assessing the shape of beach balls almost one kilometer away through fog”, Tamminen ponders.
The research addressed the importance of interdisciplinary know-how by combining biomedical engineering, imaging physics, data sciences, and biomaterial and stem-cell technologies. With the new optimized tools and methods, µCT devices can be used for imaging cellular targets efficiently for having immediate aid for breaking the current research challenges in tissue engineering and biomedical research.
The research was funded by Academy of Finland, Business Finland (TEKES), and Council of the Tampere region.
Tamminen, I., Lehto, K., Hannula, M. et al. A tube-source X-ray microtomography approach for quantitative 3D microscopy of optically challenging cell-cultured samples. Commun Biol 3, 548 (2020).
https://doi.org/10.1038/s42003-020-01273-w
Read the article: https://rdcu.be/b7ZXs
Watch a video related to the research. The video presents µCT-imaged biomaterial fibers on which stem-cell nuclei are antibody labeled: https://static-content.springer.com/esm/art%3A10.1038%2Fs42003-020-01273-w/MediaObjects/42003_2020_1273_MOESM5_ESM.mov
Further information: Ilmari Tamminen, ilmari.tamminen@tuni.fi