Researchers at NeuroGroup at CoEBoC have recently published two complementary studies in the field of human in vitro epilepsy modeling. The findings provide new insight into Dravet syndrome–related neuronal dysfunction and define how human cortical networks respond to kainic acid (KA), a widely used convulsant in experimental seizure models. NeuroGroup’s epilepsy research has already been supported by many foundations over the years such as Research Council of Finland, EU, Business Finland (MEMO) and Novo Nordisk Foundation. Lately, Doctoral Researcher Oskari Kulta has acquired more grants from multiple foundations, including Epilepsy Research Foundation, Orion Research Foundation and Brain Research Society of Finland. The research is led by PI Adj. Prof. Susanna Narkilahti at the Faculty of Medicine and Health Technology (MET) at Tampere University and is conducted with multiple international collaborators.

Dravet syndrome study reveals variant-specific network phenotypes
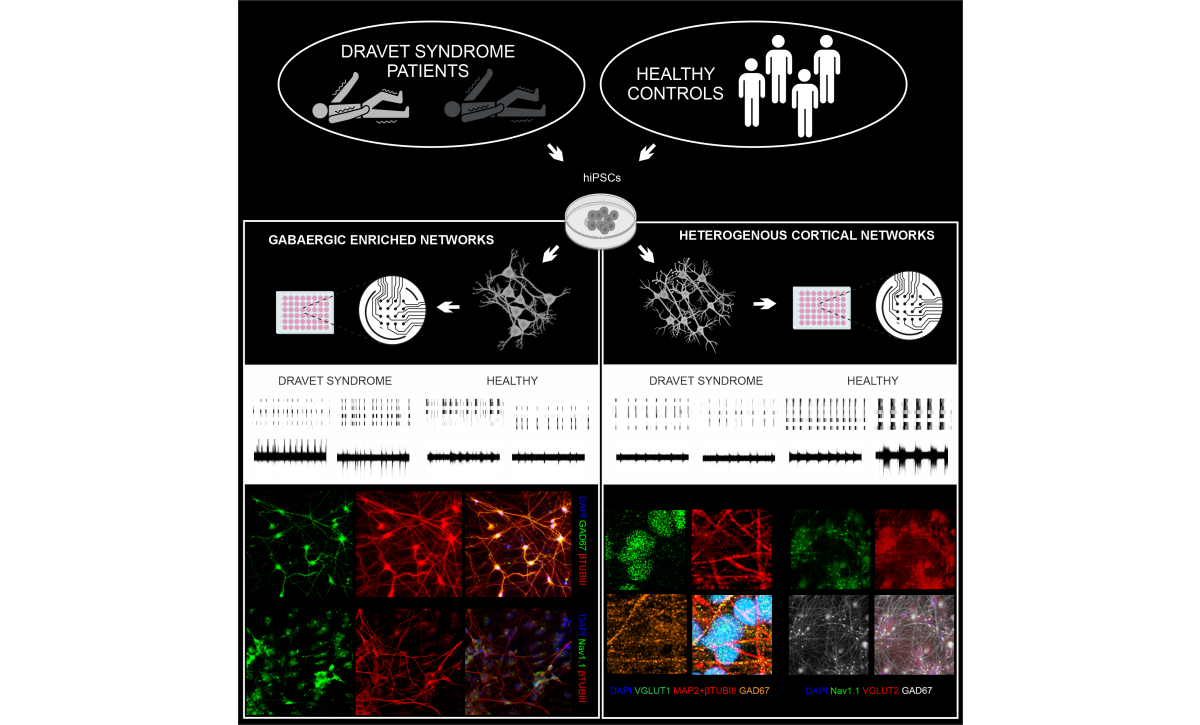
Dravet syndrome is a severe, genetic childhood epilepsy caused by mutations in SCN1A gene. The study Abnormalities in the functional activity of neural networks in a human iPSC model of Dravet syndrome (Neuroscience Research, 9/2025) authored by Ropafadzo Mzezewa, Tanja Hyvärinen, Oskari Kulta and colleagues, investigated how different SCN1A pathogenic variants alter the function of hiPSC -derived neuronal networks. Using microelectrode arrays and both GABAergic and mixed excitatory–inhibitory cortical neurons, the researchers compared patient-derived and control networks over long-term development. Interestingly, functional alterations in neuronal network activity reflected the clinical severity of the underlying SCN1A variants. Heterogeneous cortical cultures were particularly sensitive in detecting these differences, demonstrating their relevance for modeling Dravet syndrome -related network dysfunction. By providing a controlled platform for assessing patient-specific functional phenotypes, the study supports future efforts toward more personalized approaches to genetic epilepsies.
Defining kainic acid–induced responses in human cortical neurons
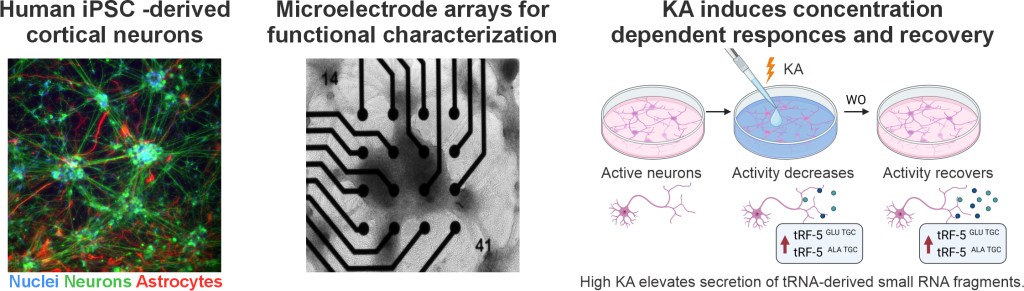
The second publication, In vitro identification of kainic acid-induced, concentration-dependent responses in human cortical neuronal networks (Neuroscience 11/2025) maps how human cortical neurons react to increasing concentrations of kainic acid (KA), a compound widely used to induce seizures and seizure-like activity in experimental models. The study conducted by Lotta Isosaari, Oskari Kulta, Satu Jäntti and colleagues shows that KA produces dose-dependent, reversible changes in neuronal firing and bursting without causing cytotoxicity. At higher concentrations, KA also triggers a measurable stress and seizure-related increase in tRNA-derived fragments, offering a potential molecular indicator of seizure-like activity. By providing a well-characterized human response profile, the work strengthens the relevance of KA-based in vitro models and supports better translation between animal studies and human epilepsy research.
International outreach and collaboration
Over the years, both published works have been presented in many field-relevant conferences such as European Dravet Syndrome Conferences 2024-2025 (Madrid, Spain), 15th European Epilepsy Congress (Rome, Italy), FENS 2024 (Vienna, Austria), Neuroscience Finland 2025 (Helsinki, Finland) and more. The existing collaboration with e.g., Uppsala University, Sweden and Royal College of Surgeons, Ireland are further supported by new exiting joint ventures from around Europe.
Future directions:
Epilepsy affects over 65 million people, and more than a third of patients do not respond to current treatments. While animal models remain essential, their ability to predict human outcomes is limited. Human stem cell–based and organ-on-chip models offer greater human relevance and can capture patient-specific seizure mechanisms, providing valuable tools for developing more effective and personalized therapies. Thus, the next step is to advance hiPSC-based epilepsy and seizure models on chip by applying a multimodal approach: combining genome-wide profiling with functional readouts to achieve more accurate disease modeling and improved support for drug development.