Modelling the Morphology and Function of Astrocytes
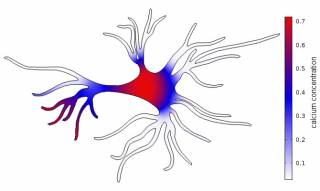
Neural cell networks derived from human pluripotent stem cells provide an excellent tool for basic research, e.g. on development of human neural systems, but as well warranted methods for toxicology and drug studies. We use computational modelling of single cell astrocytes and neuron-astrocyte networks to enhance our understanding of the astrocyte’s role in the brain. Here, we currently focus on glutamate as a gliotransmitter, potassium and the second messenger calcium. On other aspect is to understand the relationship between the astrocyte’s morphology and the internal calcium dynamics. In the long run, we aim to gain knowledge about the impact of astrocytes in neurodegenerative diseases like Alzheimer’s and epilepsy.
We are also conducting measurement on neural networks.
Understanding hIPSC Cardiomyocytes
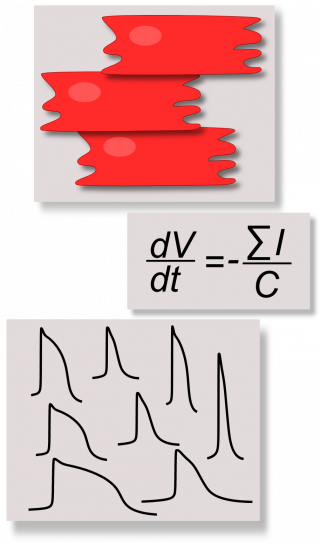
We develop computational models to discover new insights on the electrophysiology of control and diseased cardiomyocytes derived from human induced pluripotent stem cells (hiPSC-CMs) and new in silico tools to reduce the wet lab experiment time and costs, in order to assess the real usefulness of hiPSC-CMs as tools for drug testing. hiPSCs are stem cells produced by reprogramming patient somatic cells in a way they present the most common pluripotency markers and they can be differentiatied into a variety of specific phenotype, included cardiac cells. Since hiPSC-CMs have been discovered in 2007, they have been considered immediately promising in vitro tools for regenerative medicine, aimed to overcome the main issues related to the use of embryonic stem cells such as ethical problems and rejection risks. In addition, their property of reproducing in vitro the same pathological behavior of the donor patient triggered the interest in using hiPSC-CMs as drug screening/testing tools.
Recently, we started using the “Population of in silico models” approach, combined to our hiPSC-CM models, to capture the variability of the hiPSC-CM experimental data and to provide our biologists more realistic simulations.
Selected Publications
- Paci, et al. Automatic Optimization of an in Silico Model of Human iPSC Derived Cardiomyocytes Recapitulating Calcium Handling Abnormalities. Front Physiol. 2018;9:709. [PubMed]
- Paci, et al. Phenotypic variability in LQT3 human induced pluripotent stem cell-derived cardiomyocytes and their response to antiarrhythmic pharmacologic therapy: An in silico approach. Heart Rhythm. 2017;14(11):1704-1712. [PudMed]
- Paci, et al. Human induced pluripotent stem cell-derived versus adult cardiomyocytes: an in silico electrophysiological study on effects of ionic current block. Br J Pharmacol. 2015;172(21):5147-60. [PubMed]
- Paci, et al. Computational models of ventricular- and atrial-like human induced pluripotent stem cell derived cardiomyocytes. Ann Biomed Eng. 2013;41(11):2334-48. [PubMed]
Mechanisms of Cardiac Arrhythmias and Contractile Dysfunction: Virtual Heart Models as Tools for Translational Research
We aim at unravelling pathophysiological mechanisms that cause and maintain arrhythmias and contractile dysfunction, for example in atrial fibrillation and cardiomyopathies. For this purpose, we will employ existing/published mathematical models of cardiac cells (hA-CM, hV-CM, hiPSC-CM, …) and tissue, as well as, develop novel models with improved physiological accuracy of electrical and mechanical function and their interaction. Computer simulations enable quantitative investigation of cardiac physiology and disease mechanisms, which offers a gateway for discovering new drug targets and developing improved therapeutic approaches in the long-term.
Modelling the Biophysical Properties of Epithelial Tissues
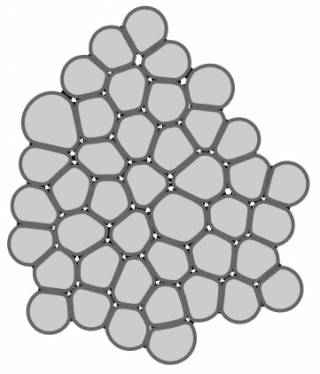
Epithelial tissues line all organs in our bodies and often reside in adverse conditions. They are thus prone to many diseases – e.g. age-related macular regeneration and inflammatory bowel syndrome – which have a huge impact on our quality of life. Our aim is to model the biophysical properties of epithelial tissues, especially their critical role as barriers and their mechanical properties. We use data from our electrophysiology measurements, calcium imaging, and confocal microscopy to develop epithelial barrier models using finite element method (FEM) and multi-compartmental approach. Further, we are developing a mechanical model of epithelial monolayer with cell-based methods to better understand the role of forces in the epithelial processes.
Selected Publications
- Tervonen et al. Structural dynamics of tight junctions modulate the properties of the epithelial barrier. PLOS One. 2019;14(4). [PubMed]
- Vainio, et al. Computational Model of Ca2+ Wave Propagation in Human Retinal Pigment Epithelial ARPE-19 Cells. PLOS One. 2015;10(6). [PubMed]
- Tervonen, et al. Prediction of passive drug permeability across the blood-retinal barrier. Pharm Res. 2014;31(9):2297-2311. [PudMed]